This study of the Galton-Watson Process, a branching process, solves problems around the development of a population under certain conditions. Galton originally proposed a mathematical question regarding the distribution of surnames in an idealized population.
Let (Zn)n≥0 be a sequence of integer-valued random variables defined by
\Zetan=∑Znk=1Xn,k,n≥1
where Xn,k:n,k∈\N forms a family of of independent and identically distributed natural-valued random variables.
The extinction probablity is given by
limPi(Zn=0)
import matplotlib.pyplot as plt
from scipy.stats import poisson
from functools import reduce
from numpy.polynomial import Polynomial
def compose(fn, x, n):
return reduce(lambda res, func: func(res), [fn] * n, x)
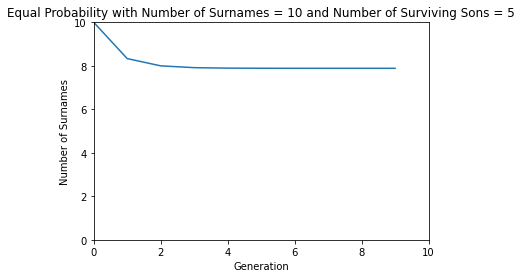
def equal_probability():
number_of_surnames = 10
number_of_surviving_sons = 5
number_of_generations = 10
fn = Polynomial([1 / (number_of_surviving_sons + 1)] * (number_of_surviving_sons + 1))
plot_graph(fn,
number_of_surnames,
number_of_generations,
"Equal Probability with Number of Surnames = {0} and Number of Surviving Sons = {1}".format(number_of_surnames, number_of_surviving_sons))
def varying_likelihood():
number_of_surnames = 10
number_of_surviving_sons = 5
number_of_generations = 10
chance_of_i_surviving_sons = [1 / (number_of_surviving_sons + i) for i in range(1, number_of_surviving_sons + 1)]
chance_of_0_surviving_sons = 1 - sum(chance_of_i_surviving_sons)
fn = Polynomial([chance_of_0_surviving_sons] + chance_of_i_surviving_sons)
plot_graph(fn,
number_of_surnames,
number_of_generations,
"Varying Likelyhood with Number of Surnames = {0} and Number of Surviving Sons = {1}".format(
number_of_surnames, number_of_surviving_sons)
)
def poisson_distribution():
number_of_surnames = 10
number_of_surviving_sons = 5
mean_number_of_surviving_sons = 1
number_of_generations = 10
chance_of_i_surviving_sons = [
poisson.pmf(i, mean_number_of_surviving_sons) for i in range(0, number_of_surviving_sons + 1)
]
fn = Polynomial(chance_of_i_surviving_sons)
plot_graph(fn,
number_of_surnames,
number_of_generations,
"Poisson Distribution with Number of Surnames = {0} and Number of Surviving Sons = {1}".format(
number_of_surnames, number_of_surviving_sons)
)
def plot_graph(fn, number_of_surnames, number_of_generations, title):
generating_functions = [compose(fn, 0, generation) for generation in range(0, number_of_generations)]
points = [number_of_surnames - number_of_surnames * func for func in generating_functions]
plt.plot(points)
plt.xlabel('Generation')
plt.ylabel('Number of Surnames')
plt.title(title)
plt.axis([0, number_of_generations, 0, number_of_surnames])
plt.show()
equal_probability()
varying_likelihood()
poisson_distribution()

Let's simulate South Korean surnames to see which surnames are likely to die out in nth generation.
References: